In this vignette we briefly illustrate how to fit PAMMs for left-truncated data. As an example we use the data already discussed in the left-truncation section of the data transformation vignette.
We first we load some libraries as well as the data.
library(eha)
library(pammtools)
library(ggplot2)
theme_set(theme_bw())
data(infants, package = "eha")
head(infants)## stratum enter exit event mother age sex parish civst ses year
## 1 1 55 365 0 dead 26 boy Nedertornea married farmer 1877
## 2 1 55 365 0 alive 26 boy Nedertornea married farmer 1870
## 3 1 55 365 0 alive 26 boy Nedertornea married farmer 1882
## 4 2 13 76 1 dead 23 girl Nedertornea married other 1847
## 5 2 13 365 0 alive 23 girl Nedertornea married other 1847
## 6 2 13 365 0 alive 23 girl Nedertornea married other 1848The header shows that we have information on entry
(enter) and exit (exit) times into the
riskset. Survival is indicated by the event variable. The
variable of interest is mother, i.e., whether mother was
alive or dead. For each child who’s mother died, 2 children of the same
age and same values for the other covariates were included into the
study who’s mother was still alive at inclusion age. The maximum follow
up was 365 days.
As a proof of concept we show that we can recreate the effects found
by the Cox model for left truncated data as implemented in
eha::coxreg:
# fit cox model for left-truncated data
fit <- coxreg(
formula = Surv(enter, exit, event) ~ mother,
data = infants)
# fit pam to left-truncated data
infants_ped <- infants %>% as_ped(Surv(enter, exit, event)~.)
pam <- pamm(
formula = ped_status ~ s(tend)+ mother,
trafo_args = list(formula = Surv(enter, exit, event)~.),
data = infants)
# compare coefficients
cbind(coef(pam)[2], coef(fit))## [,1] [,2]
## motherdead 1.68071 1.69185The coefficients of the mother variable are very similar
and indicate that children with diseased mother’s are about 5.37 times
as likely to die within one year after birth.
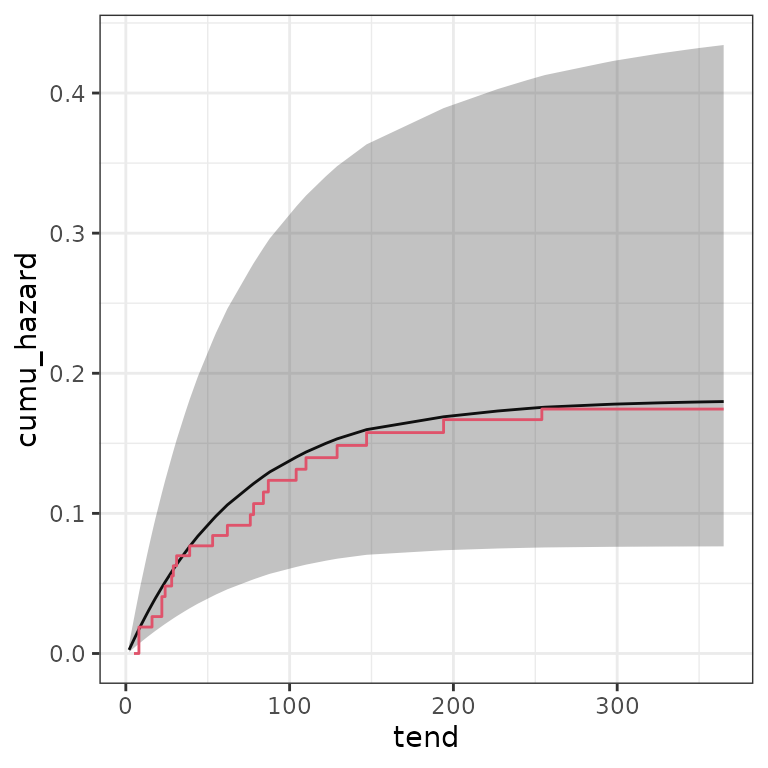
## compare (baseline) cumulative hazard probabilities
base <- survival::basehaz(fit)
ndf <- make_newdata(infants_ped, tend = unique(tend), mother = c("alive")) %>%
add_cumu_hazard(pam)
ggplot(ndf, aes(x = tend, y = cumu_hazard)) +
geom_line() +
geom_ribbon(aes(ymin = cumu_lower, ymax = cumu_upper), alpha = .3) +
geom_step(data = base, aes(x = time, y = hazard), col = 2)