library(dplyr)
library(ggplot2)
theme_set(theme_bw())
library(mgcv)
library(survival)
library(pammtools)
Set1 <- RColorBrewer::brewer.pal(9, "Set1")Stratified baselines
In some cases the proportional hazards assumption for different groups (levels of a factor variable) is violated. One approach to resolve this problem is to fit a so called stratified Cox model, where each level of factor variable will have its own baseline-hazard:
.
For illustration we again use the veteran data (censored
at day 400) from the survival package, where the cell type
of the lung cancer had four levels:
veteran <- survival::veteran # load veteran data
veteran <- veteran %>%
mutate(
trt = 1*(trt == 2),
prior = 1*(prior==10)) %>%
filter(time < 400) # restriction for illustration
table(veteran$celltype)##
## squamous smallcell adeno large
## 30 48 27 26Stratified Cox model
Fitting a stratified Cox model using the coxph function
from the survival package is simple, including a
strata term in the model formula.
The different baselines are visualized below:
baseline_gg <- ggplot(base, aes(x=time)) +
geom_step(aes(y=hazard, group=strata)) +
ylab(expression(hat(Lambda)(t))) + xlab("t")
baseline_gg + aes(col=strata)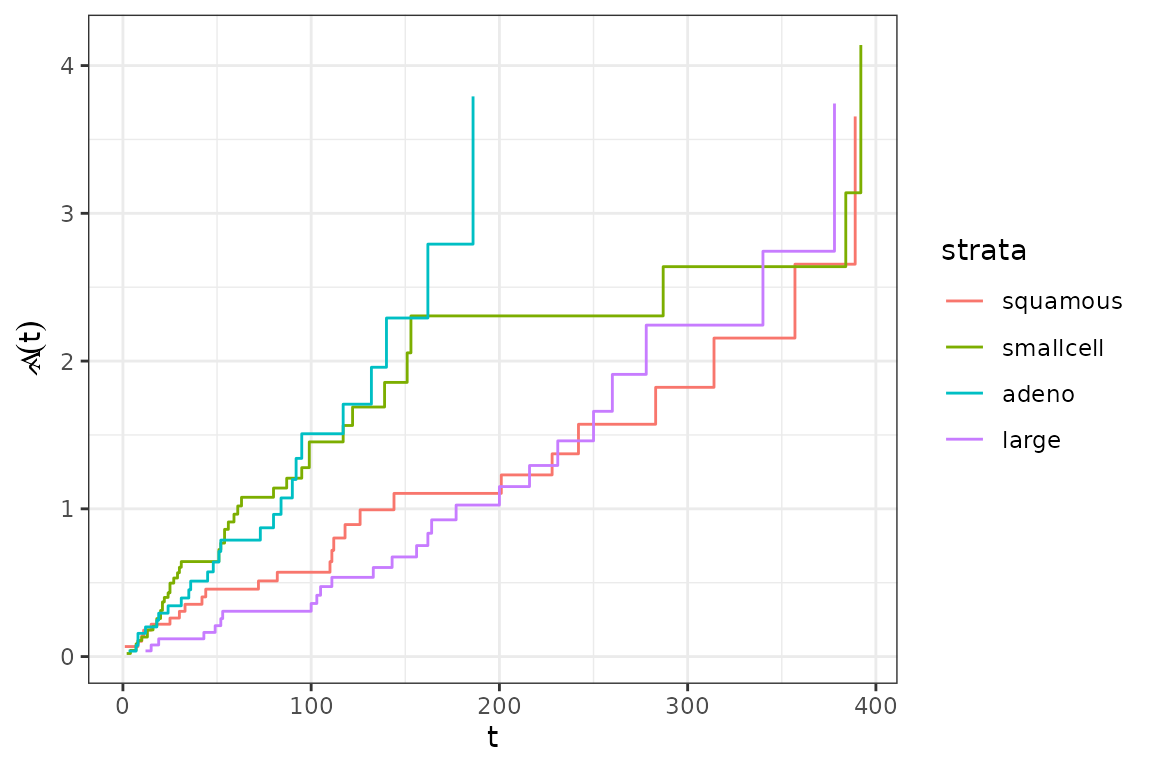
Stratified PAM
We can obtain similar results with PAMs by fitting one smooth
baseline estimate for each category of celltype in the
data. Technically, we specify the by argument in
mgcv s(...) function.
Note that in contrast to the model specification in
coxph we need to include the celltype factor
variable in the model specification:
ped <- veteran %>% as_ped(Surv(time, status)~., id="id")
pam <- gam(ped_status ~ celltype + s(tend, by=celltype), data=ped,
family=poisson(), offset=offset)Comparing the estimated baselines we get very similar results between the stratified Cox and stratified PAM models.
pinf <- ped %>%
group_by(celltype) %>%
ped_info() %>%
add_cumu_hazard(pam) %>%
rename(strata=celltype) %>%
filter(!(strata == "adeno" & tend > 200))
baseline_gg + aes(col="Nelson-Aalen") +
geom_line(data=pinf, aes(x=tend, y=cumu_hazard, group=strata, col="PAM")) +
geom_ribbon(data=pinf, aes(x=tend, ymin=cumu_lower, ymax = cumu_upper,
group=strata), fill = "black", col = NA, alpha = .2) +
facet_wrap(~strata, nrow=1) +
scale_color_manual(name="Method", values=c(Set1[1], "black")) +
theme(legend.position = "bottom")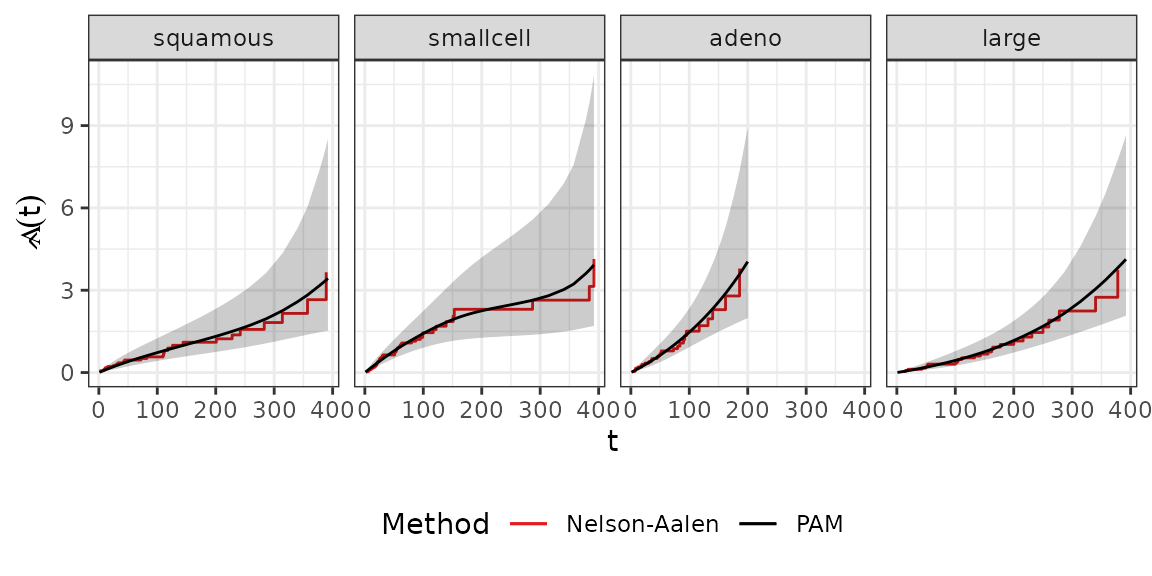
Stratified Proportional Hazards Model
Both models can be easily extended to incorporate further covariates.
If we assume that the covariate effects are not time-varying, this
extension constitutes a so called stratified proportional hazards model.
For example, including effects for treatment (trt), age
(age) and the Karnofsky-Score (karno), we get
the model
where is the baseline for observation with cell type , :
cph2 <- update(cph, .~. + trt + age + karno)
pam2 <- update(pam, .~. + trt + age + karno)
cbind(pam=coef(pam2)[5:7], cox=coef(cph2))## pam cox
## trt 0.31096123 0.32764309
## age -0.01642337 -0.01679495
## karno -0.03670427 -0.03696870Again, covariate estimates are very similar between the two models and can be interpreted in the same way.